BGGN 213 Spring 2019 Classwork
https://androidpcguy.github.io/bggn213/
Class 13: Genome Sequencing I
Akshara Balachandra 5/15/2019
MXL Genotype analysis
Read in the data….
dat <- read.csv('data/rs8067378_variation.csv', header = TRUE)
head(dat)
## Sample..Male.Female.Unknown. Genotype..forward.strand. Population.s.
## 1 NA19648 (F) A|A ALL, AMR, MXL
## 2 NA19649 (M) G|G ALL, AMR, MXL
## 3 NA19651 (F) A|A ALL, AMR, MXL
## 4 NA19652 (M) G|G ALL, AMR, MXL
## 5 NA19654 (F) G|G ALL, AMR, MXL
## 6 NA19655 (M) A|G ALL, AMR, MXL
## Father Mother
## 1 - -
## 2 - -
## 3 - -
## 4 - -
## 5 - -
## 6 - -
gen.bad <- 'G|G'
num.asthma.homoz <- sum(dat$Genotype..forward.strand. == gen.bad)
#### OR
table(dat$Genotype..forward.strand.)
##
## A|A A|G G|A G|G
## 22 21 12 9
Q5: What proportion of the Mexican Ancestry in Los Angeles sample population (MXL) are homozygous for the asthma associated SNP (G|G)?
14.1% of MXL population is homozygous for the asthma associated SNP.
Initial RNA-seq analysis
Quality score analysis
library(seqinr)
library(gtools)
phread <- asc(s2c('DDDDCDEDCDDDBBDDDCC@')) - 33
phread
## D D D D C D E D C D D D B B D D D C C @
## 35 35 35 35 34 35 36 35 34 35 35 35 33 33 35 35 35 34 34 31
Expression analysis
data <- read.csv('data/rs8067378_ENSG00000172057.6.txt', sep = ' ', header = TRUE)
head(data)
## sample geno exp
## 1 HG00367 A/G 28.96038
## 2 NA20768 A/G 20.24449
## 3 HG00361 A/A 31.32628
## 4 HG00135 A/A 34.11169
## 5 NA18870 G/G 18.25141
## 6 NA11993 A/A 32.89721
print(summary(data))
## sample geno exp
## HG00096: 1 A/A:108 Min. : 6.675
## HG00097: 1 A/G:233 1st Qu.:20.004
## HG00099: 1 G/G:121 Median :25.116
## HG00100: 1 Mean :25.640
## HG00101: 1 3rd Qu.:30.779
## HG00102: 1 Max. :51.518
## (Other):456
print(table(data$geno))
##
## A/A A/G G/G
## 108 233 121
#library(dplyr)
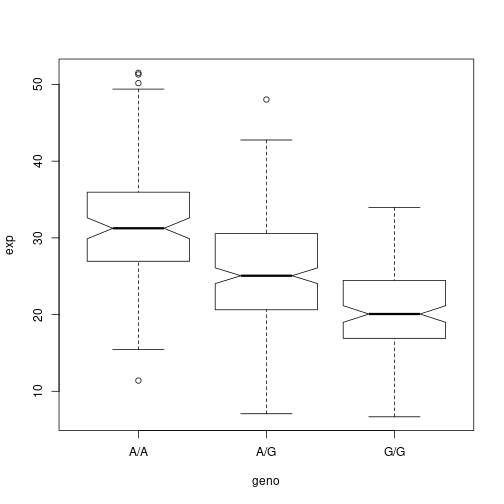
summary(data[data$geno == 'G/G',]$exp)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 6.675 16.903 20.074 20.594 24.457 33.956
summary(data[data$geno == 'A/G',]$exp)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 7.075 20.626 25.065 25.397 30.552 48.034
summary(data[data$geno == 'A/A',]$exp)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 11.40 27.02 31.25 31.82 35.92 51.52
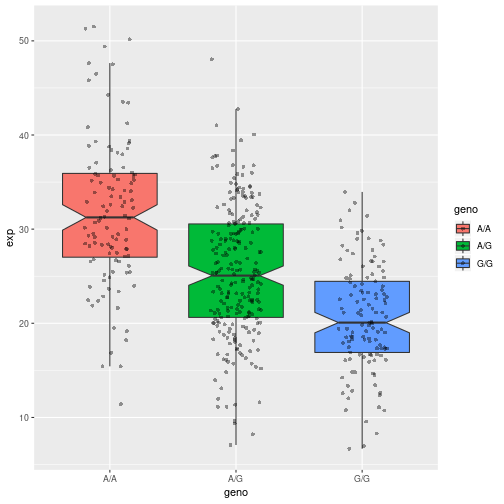
boxplot(exp ~ geno, data, notch = T)
ggplot plots because they’re prettier…
library(ggplot2)
## Registered S3 methods overwritten by 'ggplot2':
## method from
## [.quosures rlang
## c.quosures rlang
## print.quosures rlang
# Boxplot with the data shown
ggplot(data, aes(geno, exp, fill=geno)) +
geom_boxplot(notch=TRUE, outlier.shape = NA) +
geom_jitter(shape=16, position=position_jitter(0.2), alpha=0.4)