BGGN 213 Spring 2019 Classwork
https://androidpcguy.github.io/bggn213/
Class 15: Pathway analysis from RNA-seq results
Akshara Balachandra 05/22/19
Differential Expression analysis
library(DESeq2)
## Loading required package: S4Vectors
## Loading required package: stats4
## Loading required package: BiocGenerics
## Loading required package: parallel
##
## Attaching package: 'BiocGenerics'
## The following objects are masked from 'package:parallel':
##
## clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
## clusterExport, clusterMap, parApply, parCapply, parLapply,
## parLapplyLB, parRapply, parSapply, parSapplyLB
## The following objects are masked from 'package:stats':
##
## IQR, mad, sd, var, xtabs
## The following objects are masked from 'package:base':
##
## anyDuplicated, append, as.data.frame, basename, cbind,
## colnames, dirname, do.call, duplicated, eval, evalq, Filter,
## Find, get, grep, grepl, intersect, is.unsorted, lapply, Map,
## mapply, match, mget, order, paste, pmax, pmax.int, pmin,
## pmin.int, Position, rank, rbind, Reduce, rownames, sapply,
## setdiff, sort, table, tapply, union, unique, unsplit, which,
## which.max, which.min
##
## Attaching package: 'S4Vectors'
## The following object is masked from 'package:base':
##
## expand.grid
## Loading required package: IRanges
## Loading required package: GenomicRanges
## Loading required package: GenomeInfoDb
## Loading required package: SummarizedExperiment
## Loading required package: Biobase
## Welcome to Bioconductor
##
## Vignettes contain introductory material; view with
## 'browseVignettes()'. To cite Bioconductor, see
## 'citation("Biobase")', and for packages 'citation("pkgname")'.
## Loading required package: DelayedArray
## Loading required package: matrixStats
##
## Attaching package: 'matrixStats'
## The following objects are masked from 'package:Biobase':
##
## anyMissing, rowMedians
## Loading required package: BiocParallel
##
## Attaching package: 'DelayedArray'
## The following objects are masked from 'package:matrixStats':
##
## colMaxs, colMins, colRanges, rowMaxs, rowMins, rowRanges
## The following objects are masked from 'package:base':
##
## aperm, apply, rowsum
## Registered S3 methods overwritten by 'ggplot2':
## method from
## [.quosures rlang
## c.quosures rlang
## print.quosures rlang
library(dplyr)
##
## Attaching package: 'dplyr'
## The following object is masked from 'package:matrixStats':
##
## count
## The following object is masked from 'package:Biobase':
##
## combine
## The following objects are masked from 'package:GenomicRanges':
##
## intersect, setdiff, union
## The following object is masked from 'package:GenomeInfoDb':
##
## intersect
## The following objects are masked from 'package:IRanges':
##
## collapse, desc, intersect, setdiff, slice, union
## The following objects are masked from 'package:S4Vectors':
##
## first, intersect, rename, setdiff, setequal, union
## The following objects are masked from 'package:BiocGenerics':
##
## combine, intersect, setdiff, union
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
Load the data…
metadata <- read.csv('data/GSE37704_metadata.csv', row.names = 1)
counts <- read.csv('data/GSE37704_featurecounts.csv', row.names = 1)
head(metadata)
## condition
## SRR493366 control_sirna
## SRR493367 control_sirna
## SRR493368 control_sirna
## SRR493369 hoxa1_kd
## SRR493370 hoxa1_kd
## SRR493371 hoxa1_kd
head(counts)
## length SRR493366 SRR493367 SRR493368 SRR493369 SRR493370
## ENSG00000186092 918 0 0 0 0 0
## ENSG00000279928 718 0 0 0 0 0
## ENSG00000279457 1982 23 28 29 29 28
## ENSG00000278566 939 0 0 0 0 0
## ENSG00000273547 939 0 0 0 0 0
## ENSG00000187634 3214 124 123 205 207 212
## SRR493371
## ENSG00000186092 0
## ENSG00000279928 0
## ENSG00000279457 46
## ENSG00000278566 0
## ENSG00000273547 0
## ENSG00000187634 258
Remove the first column from counts dataframe.
counts <- as.matrix(counts[,2:ncol(counts)])
head(counts)
## SRR493366 SRR493367 SRR493368 SRR493369 SRR493370
## ENSG00000186092 0 0 0 0 0
## ENSG00000279928 0 0 0 0 0
## ENSG00000279457 23 28 29 29 28
## ENSG00000278566 0 0 0 0 0
## ENSG00000273547 0 0 0 0 0
## ENSG00000187634 124 123 205 207 212
## SRR493371
## ENSG00000186092 0
## ENSG00000279928 0
## ENSG00000279457 46
## ENSG00000278566 0
## ENSG00000273547 0
## ENSG00000187634 258
Filter out all rows with 0 values
filteredCounts <- counts[rowSums(counts) != 0,]
head(filteredCounts)
## SRR493366 SRR493367 SRR493368 SRR493369 SRR493370
## ENSG00000279457 23 28 29 29 28
## ENSG00000187634 124 123 205 207 212
## ENSG00000188976 1637 1831 2383 1226 1326
## ENSG00000187961 120 153 180 236 255
## ENSG00000187583 24 48 65 44 48
## ENSG00000187642 4 9 16 14 16
## SRR493371
## ENSG00000279457 46
## ENSG00000187634 258
## ENSG00000188976 1504
## ENSG00000187961 357
## ENSG00000187583 64
## ENSG00000187642 16
Running DEseq2
dds <- DESeqDataSetFromMatrix(countData = filteredCounts, colData = metadata,
design= ~condition)
dds <- DESeq(dds)
## estimating size factors
## estimating dispersions
## gene-wise dispersion estimates
## mean-dispersion relationship
## final dispersion estimates
## fitting model and testing
dds
## class: DESeqDataSet
## dim: 15975 6
## metadata(1): version
## assays(4): counts mu H cooks
## rownames(15975): ENSG00000279457 ENSG00000187634 ...
## ENSG00000276345 ENSG00000271254
## rowData names(22): baseMean baseVar ... deviance maxCooks
## colnames(6): SRR493366 SRR493367 ... SRR493370 SRR493371
## colData names(2): condition sizeFactor
Extract the results…
res <- results(dds)
summary(res)
##
## out of 15975 with nonzero total read count
## adjusted p-value < 0.1
## LFC > 0 (up) : 4349, 27%
## LFC < 0 (down) : 4396, 28%
## outliers [1] : 0, 0%
## low counts [2] : 1237, 7.7%
## (mean count < 0)
## [1] see 'cooksCutoff' argument of ?results
## [2] see 'independentFiltering' argument of ?results
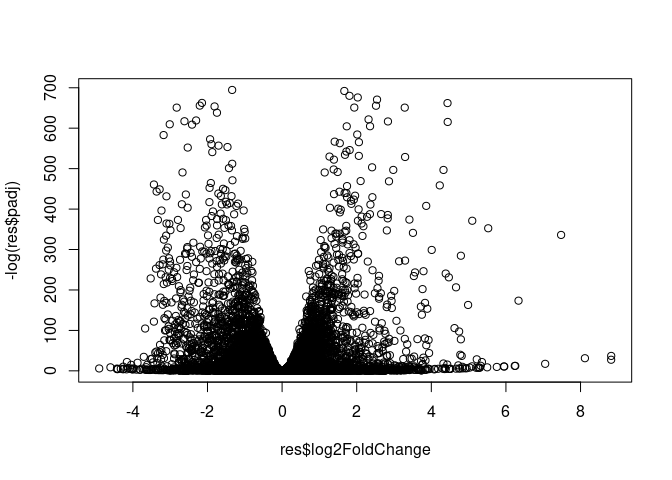
Volcano Plot
plot(res$log2FoldChange, -log(res$padj))
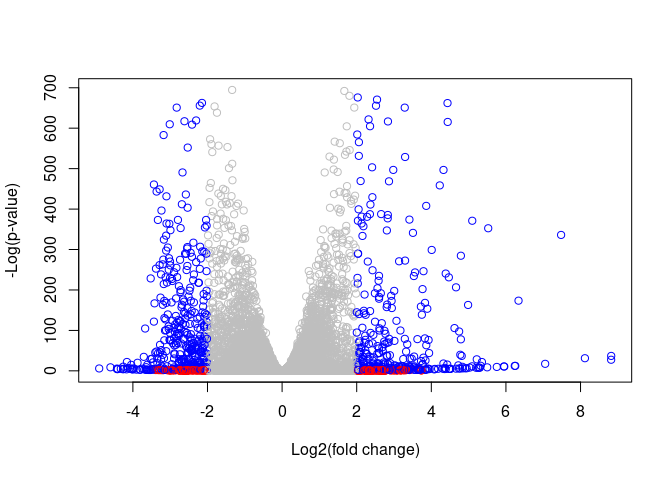
Adding all the colors!!!!
mycol <- rep('gray', nrow(res))
mycol[abs(res$log2FoldChange) > 2] <- 'red'
mycol[(abs(res$log2FoldChange) > 2) & (-log(res$padj) > 2)] <- 'blue'
plot(res$log2FoldChange, -log(res$padj), col = mycol,
xlab = "Log2(fold change)", ylab = '-Log(p-value)')
nrow(filteredCounts)
## [1] 15975
Add gene annotation
library(AnnotationDbi)
##
## Attaching package: 'AnnotationDbi'
## The following object is masked from 'package:dplyr':
##
## select
library(org.Hs.eg.db)
##
#columns(org.Hs.eg.db)
res$symbol = mapIds(org.Hs.eg.db,
keys = row.names(res),
keytype="ENSEMBL",
column='SYMBOL',
multiVals="first")
## 'select()' returned 1:many mapping between keys and columns
res$entrez = mapIds(org.Hs.eg.db,
keys=row.names(res),
keytype="ENSEMBL",
column="ENTREZID",
multiVals="first")
## 'select()' returned 1:many mapping between keys and columns
res$name = mapIds(org.Hs.eg.db,
keys=row.names(res),
keytype='ENSEMBL',
column='GENENAME',
multiVals="first")
## 'select()' returned 1:many mapping between keys and columns
head(res, 10)
## log2 fold change (MLE): condition hoxa1 kd vs control sirna
## Wald test p-value: condition hoxa1 kd vs control sirna
## DataFrame with 10 rows and 9 columns
## baseMean log2FoldChange lfcSE
## <numeric> <numeric> <numeric>
## ENSG00000279457 29.9135794276176 0.17925708367269 0.324821565250144
## ENSG00000187634 183.229649921658 0.426457118403308 0.140265820376891
## ENSG00000188976 1651.18807619944 -0.692720464846376 0.0548465415913904
## ENSG00000187961 209.637938486147 0.729755610585223 0.131859899969346
## ENSG00000187583 47.2551232589398 0.0405765278756312 0.271892808601773
## ENSG00000187642 11.9797501642461 0.542810491577361 0.521559849534146
## ENSG00000188290 108.922127976716 2.0570638345631 0.196905312993834
## ENSG00000187608 350.71686801731 0.257383686481769 0.102726560033536
## ENSG00000188157 9128.439421961 0.389908792022773 0.0467163395511382
## ENSG00000237330 0.158192358990472 0.785955208142751 4.0804728567969
## stat pvalue
## <numeric> <numeric>
## ENSG00000279457 0.551863246932651 0.58104205074703
## ENSG00000187634 3.04034951107424 0.00236303749730974
## ENSG00000188976 -12.6301576133493 1.4398954015447e-36
## ENSG00000187961 5.53432552849555 3.12428248077812e-08
## ENSG00000187583 0.149237223611388 0.881366448669147
## ENSG00000187642 1.04074439790984 0.297994191720985
## ENSG00000188290 10.4469696794191 1.51281875407224e-25
## ENSG00000187608 2.50552229528317 0.0122270689409813
## ENSG00000188157 8.34630443585929 7.04321148758761e-17
## ENSG00000237330 0.192613757210411 0.847261469988086
## padj symbol entrez
## <numeric> <character> <character>
## ENSG00000279457 0.686554777832897 NA NA
## ENSG00000187634 0.00515718149494313 SAMD11 148398
## ENSG00000188976 1.7654890539073e-35 NOC2L 26155
## ENSG00000187961 1.13412993107655e-07 KLHL17 339451
## ENSG00000187583 0.91903061557138 PLEKHN1 84069
## ENSG00000187642 0.403379309754088 PERM1 84808
## ENSG00000188290 1.30538189681011e-24 HES4 57801
## ENSG00000187608 0.023745228890787 ISG15 9636
## ENSG00000188157 4.21962808553115e-16 AGRN 375790
## ENSG00000237330 NA RNF223 401934
## name
## <character>
## ENSG00000279457 NA
## ENSG00000187634 sterile alpha motif domain containing 11
## ENSG00000188976 NOC2 like nucleolar associated transcriptional repressor
## ENSG00000187961 kelch like family member 17
## ENSG00000187583 pleckstrin homology domain containing N1
## ENSG00000187642 PPARGC1 and ESRR induced regulator, muscle 1
## ENSG00000188290 hes family bHLH transcription factor 4
## ENSG00000187608 ISG15 ubiquitin like modifier
## ENSG00000188157 agrin
## ENSG00000237330 ring finger protein 223
Order the results by p-value and save to disk.
res <- res[order(res$pvalue),]
write.csv(res, file = 'data/deseq_results.csv')
Pathway Analysis
Import all the packages…
library(gage)
library(gageData)
library(pathview)
## ##############################################################################
## Pathview is an open source software package distributed under GNU General
## Public License version 3 (GPLv3). Details of GPLv3 is available at
## http://www.gnu.org/licenses/gpl-3.0.html. Particullary, users are required to
## formally cite the original Pathview paper (not just mention it) in publications
## or products. For details, do citation("pathview") within R.
##
## The pathview downloads and uses KEGG data. Non-academic uses may require a KEGG
## license agreement (details at http://www.kegg.jp/kegg/legal.html).
## ##############################################################################
data(kegg.sets.hs)
data(sigmet.idx.hs)
Inspecting some of the data
# Focus on signaling and metabolic pathways only
kegg.sets.hs <- kegg.sets.hs[sigmet.idx.hs]
# Examine the first 3 pathways
head(kegg.sets.hs, 3)
## $`hsa00232 Caffeine metabolism`
## [1] "10" "1544" "1548" "1549" "1553" "7498" "9"
##
## $`hsa00983 Drug metabolism - other enzymes`
## [1] "10" "1066" "10720" "10941" "151531" "1548" "1549"
## [8] "1551" "1553" "1576" "1577" "1806" "1807" "1890"
## [15] "221223" "2990" "3251" "3614" "3615" "3704" "51733"
## [22] "54490" "54575" "54576" "54577" "54578" "54579" "54600"
## [29] "54657" "54658" "54659" "54963" "574537" "64816" "7083"
## [36] "7084" "7172" "7363" "7364" "7365" "7366" "7367"
## [43] "7371" "7372" "7378" "7498" "79799" "83549" "8824"
## [50] "8833" "9" "978"
##
## $`hsa00230 Purine metabolism`
## [1] "100" "10201" "10606" "10621" "10622" "10623" "107"
## [8] "10714" "108" "10846" "109" "111" "11128" "11164"
## [15] "112" "113" "114" "115" "122481" "122622" "124583"
## [22] "132" "158" "159" "1633" "171568" "1716" "196883"
## [29] "203" "204" "205" "221823" "2272" "22978" "23649"
## [36] "246721" "25885" "2618" "26289" "270" "271" "27115"
## [43] "272" "2766" "2977" "2982" "2983" "2984" "2986"
## [50] "2987" "29922" "3000" "30833" "30834" "318" "3251"
## [57] "353" "3614" "3615" "3704" "377841" "471" "4830"
## [64] "4831" "4832" "4833" "4860" "4881" "4882" "4907"
## [71] "50484" "50940" "51082" "51251" "51292" "5136" "5137"
## [78] "5138" "5139" "5140" "5141" "5142" "5143" "5144"
## [85] "5145" "5146" "5147" "5148" "5149" "5150" "5151"
## [92] "5152" "5153" "5158" "5167" "5169" "51728" "5198"
## [99] "5236" "5313" "5315" "53343" "54107" "5422" "5424"
## [106] "5425" "5426" "5427" "5430" "5431" "5432" "5433"
## [113] "5434" "5435" "5436" "5437" "5438" "5439" "5440"
## [120] "5441" "5471" "548644" "55276" "5557" "5558" "55703"
## [127] "55811" "55821" "5631" "5634" "56655" "56953" "56985"
## [134] "57804" "58497" "6240" "6241" "64425" "646625" "654364"
## [141] "661" "7498" "8382" "84172" "84265" "84284" "84618"
## [148] "8622" "8654" "87178" "8833" "9060" "9061" "93034"
## [155] "953" "9533" "954" "955" "956" "957" "9583"
## [162] "9615"
foldchanges <- res$log2FoldChange
names(foldchanges) <- res$entrez
head(foldchanges)
## 1266 54855 1465 51232 2034 2317
## -2.422719 3.201955 -2.313738 -2.059631 -1.888019 -1.649792
Gage pathway analyssi…..
# Get the results
keggres <- gage(foldchanges, gsets=kegg.sets.hs)
attributes(keggres)
## $names
## [1] "greater" "less" "stats"
head(keggres$less)
## p.geomean stat.mean p.val
## hsa04110 Cell cycle 8.995727e-06 -4.378644 8.995727e-06
## hsa03030 DNA replication 9.424076e-05 -3.951803 9.424076e-05
## hsa03013 RNA transport 1.375901e-03 -3.028500 1.375901e-03
## hsa03440 Homologous recombination 3.066756e-03 -2.852899 3.066756e-03
## hsa04114 Oocyte meiosis 3.784520e-03 -2.698128 3.784520e-03
## hsa00010 Glycolysis / Gluconeogenesis 8.961413e-03 -2.405398 8.961413e-03
## q.val set.size exp1
## hsa04110 Cell cycle 0.001448312 121 8.995727e-06
## hsa03030 DNA replication 0.007586381 36 9.424076e-05
## hsa03013 RNA transport 0.073840037 144 1.375901e-03
## hsa03440 Homologous recombination 0.121861535 28 3.066756e-03
## hsa04114 Oocyte meiosis 0.121861535 102 3.784520e-03
## hsa00010 Glycolysis / Gluconeogenesis 0.212222694 53 8.961413e-03
Pathway visualization
pathview(gene.data=foldchanges, pathway.id="hsa04110")
## 'select()' returned 1:1 mapping between keys and columns
## Info: Working in directory /home/aksharab/Documents/bggn213/src/class15
## Info: Writing image file hsa04110.pathview.png
Do same for top 5 downregulated…
SPECIES = 'hsa'
down5 <- unlist(strsplit(rownames(keggres$less)[1:5], split = ' '))
down5 <- down5[grepl(pattern = SPECIES, down5)]
up5 <- unlist(strsplit(rownames(keggres$greater)[1:5], split = ' '))
up5 <- up5[grepl(pattern = SPECIES, up5)]
pathview(gene.data = foldchanges, pathway.id = down5, species = SPECIES)
## 'select()' returned 1:1 mapping between keys and columns
## Info: Working in directory /home/aksharab/Documents/bggn213/src/class15
## Info: Writing image file hsa04110.pathview.png
## 'select()' returned 1:1 mapping between keys and columns
## Info: Working in directory /home/aksharab/Documents/bggn213/src/class15
## Info: Writing image file hsa03030.pathview.png
## 'select()' returned 1:1 mapping between keys and columns
## Info: Working in directory /home/aksharab/Documents/bggn213/src/class15
## Info: Writing image file hsa03013.pathview.png
## 'select()' returned 1:1 mapping between keys and columns
## Info: Working in directory /home/aksharab/Documents/bggn213/src/class15
## Info: Writing image file hsa03440.pathview.png
## 'select()' returned 1:1 mapping between keys and columns
## Info: Working in directory /home/aksharab/Documents/bggn213/src/class15
## Info: Writing image file hsa04114.pathview.png
pathview(gene.data = foldchanges, pathway.id = up5, species = SPECIES)
## 'select()' returned 1:1 mapping between keys and columns
## Info: Working in directory /home/aksharab/Documents/bggn213/src/class15
## Info: Writing image file hsa04640.pathview.png
## 'select()' returned 1:1 mapping between keys and columns
## Info: Working in directory /home/aksharab/Documents/bggn213/src/class15
## Info: Writing image file hsa04630.pathview.png
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## Warning in structure(x$children, class = "XMLNodeList"): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
## Consider 'structure(list(), *)' instead.
## 'select()' returned 1:1 mapping between keys and columns
## Info: Working in directory /home/aksharab/Documents/bggn213/src/class15
## Info: Writing image file hsa00140.pathview.png
## 'select()' returned 1:1 mapping between keys and columns
## Info: Working in directory /home/aksharab/Documents/bggn213/src/class15
## Info: Writing image file hsa04142.pathview.png
## Info: some node width is different from others, and hence adjusted!
## 'select()' returned 1:1 mapping between keys and columns
## Info: Working in directory /home/aksharab/Documents/bggn213/src/class15
## Info: Writing image file hsa04330.pathview.png
Gene ontology
data(go.sets.hs)
data(go.subs.hs)
# Focus on Biological Process subset of GO
gobpsets <- go.sets.hs[go.subs.hs$BP]
gobpres <- gage(foldchanges, gsets=gobpsets, same.dir=TRUE)
lapply(gobpres, head)
## $greater
## p.geomean stat.mean
## GO:0007156 homophilic cell adhesion 8.519724e-05 3.824205
## GO:0002009 morphogenesis of an epithelium 1.396681e-04 3.653886
## GO:0048729 tissue morphogenesis 1.432451e-04 3.643242
## GO:0007610 behavior 2.195494e-04 3.530241
## GO:0060562 epithelial tube morphogenesis 5.932837e-04 3.261376
## GO:0035295 tube development 5.953254e-04 3.253665
## p.val q.val set.size
## GO:0007156 homophilic cell adhesion 8.519724e-05 0.1952430 113
## GO:0002009 morphogenesis of an epithelium 1.396681e-04 0.1952430 339
## GO:0048729 tissue morphogenesis 1.432451e-04 0.1952430 424
## GO:0007610 behavior 2.195494e-04 0.2244344 427
## GO:0060562 epithelial tube morphogenesis 5.932837e-04 0.3712298 257
## GO:0035295 tube development 5.953254e-04 0.3712298 391
## exp1
## GO:0007156 homophilic cell adhesion 8.519724e-05
## GO:0002009 morphogenesis of an epithelium 1.396681e-04
## GO:0048729 tissue morphogenesis 1.432451e-04
## GO:0007610 behavior 2.195494e-04
## GO:0060562 epithelial tube morphogenesis 5.932837e-04
## GO:0035295 tube development 5.953254e-04
##
## $less
## p.geomean stat.mean
## GO:0048285 organelle fission 1.536227e-15 -8.063910
## GO:0000280 nuclear division 4.286961e-15 -7.939217
## GO:0007067 mitosis 4.286961e-15 -7.939217
## GO:0000087 M phase of mitotic cell cycle 1.169934e-14 -7.797496
## GO:0007059 chromosome segregation 2.028624e-11 -6.878340
## GO:0000236 mitotic prometaphase 1.729553e-10 -6.695966
## p.val q.val
## GO:0048285 organelle fission 1.536227e-15 5.843127e-12
## GO:0000280 nuclear division 4.286961e-15 5.843127e-12
## GO:0007067 mitosis 4.286961e-15 5.843127e-12
## GO:0000087 M phase of mitotic cell cycle 1.169934e-14 1.195965e-11
## GO:0007059 chromosome segregation 2.028624e-11 1.659009e-08
## GO:0000236 mitotic prometaphase 1.729553e-10 1.178690e-07
## set.size exp1
## GO:0048285 organelle fission 376 1.536227e-15
## GO:0000280 nuclear division 352 4.286961e-15
## GO:0007067 mitosis 352 4.286961e-15
## GO:0000087 M phase of mitotic cell cycle 362 1.169934e-14
## GO:0007059 chromosome segregation 142 2.028624e-11
## GO:0000236 mitotic prometaphase 84 1.729553e-10
##
## $stats
## stat.mean exp1
## GO:0007156 homophilic cell adhesion 3.824205 3.824205
## GO:0002009 morphogenesis of an epithelium 3.653886 3.653886
## GO:0048729 tissue morphogenesis 3.643242 3.643242
## GO:0007610 behavior 3.530241 3.530241
## GO:0060562 epithelial tube morphogenesis 3.261376 3.261376
## GO:0035295 tube development 3.253665 3.253665
Pathview for GO dataset
SPECIES = 'GO'
down5 <- unlist(strsplit(rownames(gobpres$less)[1:5], split = ' '))
down5 <- down5[grepl(pattern = SPECIES, down5)]
up5 <- unlist(strsplit(rownames(gobpres$greater)[1:5], split = ' '))
up5 <- up5[grepl(pattern = SPECIES, up5)]
down5
## [1] "GO:0048285" "GO:0000280" "GO:0007067" "GO:0000087" "GO:0007059"
#pathview(gene.data = foldchanges, pathway.id = down5, species = SPECIES)
#pathview(gene.data = foldchanges, pathway.id = up5, species = SPECIES)